应用昆虫学报 ›› 2020, Vol. 57 ›› Issue (4): 823-832.doi: 10.7679/j.issn.2095-1353.2020.084
• 昆虫抗药性专栏 • 上一篇
王传鹏1,2( ), 张帅2, 高雪珂2, 雒珺瑜2, 朱香镇2, 王丽2, 张开心2, 杨亦桦1,***(
), 张帅2, 高雪珂2, 雒珺瑜2, 朱香镇2, 王丽2, 张开心2, 杨亦桦1,***( ), 崔金杰2,***(
), 崔金杰2,***( )
)
收稿日期:2019-07-03
接受日期:2019-11-20
出版日期:2020-07-27
发布日期:2020-09-02
通讯作者:
杨亦桦,崔金杰
E-mail:wangcpzs@163.com;yhyang@njau.edu.cn;aycuijinjie@163.com
基金资助:
Chuan-Peng WANG1,2( ), Shuai ZHANG2, Xue-Ke GAO2, Jun-Yu LUO2, Xiang-Zhen ZHU2, Li WANG2, Kai-Xin ZHANG2, Yi-Hua YANG1,***(
), Shuai ZHANG2, Xue-Ke GAO2, Jun-Yu LUO2, Xiang-Zhen ZHU2, Li WANG2, Kai-Xin ZHANG2, Yi-Hua YANG1,***( ), Jin-Jie CUI2,***(
), Jin-Jie CUI2,***( )
)
Received:2019-07-03
Accepted:2019-11-20
Online:2020-07-27
Published:2020-09-02
Contact:
Yi-Hua YANG,Jin-Jie CUI
E-mail:wangcpzs@163.com;yhyang@njau.edu.cn;aycuijinjie@163.com
摘要:
【目的】明确棉蚜Aphis gossypii Glover体内3个高表达谷胱甘肽-S-转移酶(GST)基因不同发育阶段的表达谱,并比较它们在不同寄主专化型棉蚜中的表达差异。【方法】 从棉蚜的伏蚜和苗蚜转录组数据库中挑选GSTs并克隆得到全长cDNA序列,使用qRT-PCR分析这些基因在不同寄主专化型棉蚜不同龄期中的相对表达量。【结果】 克隆到3个高表达的GSTs基因,分别命名为AgoGST-s1(GenBank登录号:MN688789)、AgoGST-d1(GenBank登录号:MN688790)、AgoGST-d2(GenBank登录号:MN688791)。这3个基因在不同龄期均表达,不同发育阶段的表达谱基本一致,表达量呈现从低到高的变化趋势。AgoGST-s1基因在各个发育阶段相对表达量的变化幅度较小,而AgoGST-d1、AgoGST-d2的变化幅度较大。对比两种寄主专化型棉蚜相同龄期时的表达情况发现,黄瓜型棉蚜中GSTs基因的表达量在大部分龄期都高于棉花型棉蚜。【结论】 AgoGST-s1、AgoGST-d1、AgoGST-d2基因表达量伴随棉蚜生长发育变化趋势明显,在不同寄主专化型棉蚜体内的表达量存在差异,可能与棉蚜对寄主植物的适应性有关。
王传鹏, 张帅, 高雪珂, 雒珺瑜, 朱香镇, 王丽, 张开心, 杨亦桦, 崔金杰. 三个棉蚜谷胱甘肽-S-转移酶(GST)基因克隆及在不同寄主专化型中的表达分析 *[J]. 应用昆虫学报, 2020, 57(4): 823-832.
Chuan-Peng WANG, Shuai ZHANG, Xue-Ke GAO, Jun-Yu LUO, Xiang-Zhen ZHU, Li WANG, Kai-Xin ZHANG, Yi-Hua YANG, Jin-Jie CUI. Clone and expression analysis of three glutathione-S-transferase genes in different host-specific types of Aphis gossypii Glover[J]. Chinese Journal of Applied Entomology, 2020, 57(4): 823-832.
| 基因 Gene | 引物序列 Primer sequence(5?-3?) | 扩增效率(%) Amplification efficiency | 相关系数(R2) Correlation coefficient |
|---|---|---|---|
| AgoGST-s1 | F-TGATAATGGCACTTAACTACTA R-AACCACTATCCGAACAAC | 90 | 0.994 |
| AgoGST-d1 | F-ACAGTCAGTCGTCATACAC R-TTCTGAATCGGTCGGTTC | 91 | 0.997 |
| AgoGST-d2 | F-GTGATTGGCTTCGTTGTAAT R-ATCTGACCAGTTATCCTAATGT | 94 | 0.995 |
| AgoGAPDH | F-ATCATTCCAGCATCTACT R-TCCTTAACCTTATCCTTGA | 90 | 0.999 |
表1 实时荧光定量PCR引物
Table 1 Primers used for real-time quantitative PCR
| 基因 Gene | 引物序列 Primer sequence(5?-3?) | 扩增效率(%) Amplification efficiency | 相关系数(R2) Correlation coefficient |
|---|---|---|---|
| AgoGST-s1 | F-TGATAATGGCACTTAACTACTA R-AACCACTATCCGAACAAC | 90 | 0.994 |
| AgoGST-d1 | F-ACAGTCAGTCGTCATACAC R-TTCTGAATCGGTCGGTTC | 91 | 0.997 |
| AgoGST-d2 | F-GTGATTGGCTTCGTTGTAAT R-ATCTGACCAGTTATCCTAATGT | 94 | 0.995 |
| AgoGAPDH | F-ATCATTCCAGCATCTACT R-TCCTTAACCTTATCCTTGA | 90 | 0.999 |

图1 棉蚜AgoGST-s1与其他昆虫Sigma类GSTs氨基酸序列多重比较 AgoGST-s1:棉蚜Aphis gossypii,XP_027840459.1;BmGST-s2:家蚕Bombyx mori,NP_001036994.1;DvGST-s1:葡萄根瘤蚜Daktulosphaira vitifoliae,AUN35386.1;AmGST-s1:西方蜜蜂Apis mellifera,NP_001153742.1;LmGST-s4:飞蝗Locusta migratoria,AEB91976.1。黑色方框为AgoGST-s1 N端结构域;红色方框为AgoGST-s1 C端结构域;预测的GSH结合位点(G-site)和底物结合位点(H-site)分别标注为红色和绿色,Sigma家族特有的催化位点残基“Y8”用黑色箭头表示。
Fig. 1 Amino acid sequence alignment of AgoGST-s1 from Aphis gossypii with Sigma-class homologues from other insects Black box region is the N-terminal domain of AgoGST-s1; Red box region is the C-terminal domain of AgoGST-s1; Predicted GSH binding sites (G-sites) and substrate binding sites (H-sites) are shaded red and green, respectively. The typical catalytic site residue “Y8” of Sigma-class is indicated in black arrow.
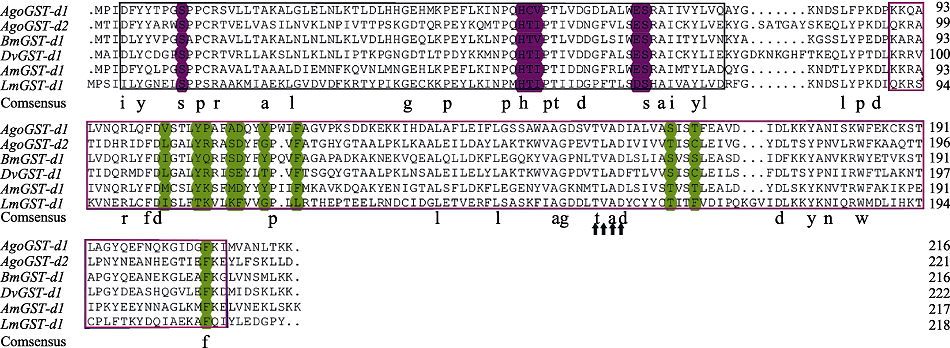
图2 棉蚜AgoGST-d1、AgoGST-d2与其他昆虫Delta类GSTs氨基酸序列多重比较 AgoGST-d1:棉蚜Aphis gossypii,AFM78644.1;AgoGST-d2:棉蚜Aphis gossypii,XP_027847501.1;BmGST-d2:家蚕Bombyx mori,NP_001036974.1;DvGST-s1:葡萄根瘤蚜Daktulosphaira vitifoliae,AUN35382.1;AmGST-d1:西方蜜蜂Apis mellifera,NP_001171499.1;LmGST-d1:飞蝗Locusta migratoria,AHC08057.1。黑色方框为AgoGST-s1 N端结构域;红色方框为AgoGST-s1 C端结构域;预测的GSH结合位点(G-site)和底物结合位点(H-site)分别标注为红色和绿色,Delta家族特有的典型序列“T*AD”用黑色箭头标注。
Fig. 2 Amino acid sequence alignment of AgoGST-d1, AgoGST-d2 from Aphis gossypii with Delta-class homologues from other insects Black box region is the N-terminal domain of AgoGST-d1, AgoGST-d2; Red box region is the C-terminal domain of AgoGST-d1, AgoGST-d2; Predicted GSH binding sites (G-sites) and substrate binding sites (H-sites) are shaded red and green, respectively. The typical sequence “T*AD” of Delta-class is marked by black arrow.

图3 棉蚜与其他不同昆虫GSTs系统进化分析 进化树使用邻接法构建,各分支置信度经由1 000次Bootstrap检验。棉蚜GST基因用红色字体表示。 So:米象;Bg:德国小蠊;Lm:东亚飞蝗;Cs:二化螟;Px:柑橘凤蝶;Aga:冈比亚按蚊;Dv:葡萄根瘤蚜;Dm:黑腹果蝇;Dp:黑脉金斑蝶;Tm:黄粉虫;Bm:家蚕;Pa:美洲大蠊;Zn:湿木白蚁;Api:豌豆蚜;Sl:斜纹夜蛾;Ls:烟草甲;Bt:烟粉虱;Ape:柞蚕;Ago:棉蚜。
Fig. 3 Phylogenetic analysis of GSTs of Aphis gossypii and other insects The tree is constructed by using neighbor-joining method. Bootstrap support values are based on 1 000 replicates. The GSTs of cotton aphid is marked with the red. So: Sitophilus oryzae; Bg: Blattella germanica; Lm: Locusta migratoria; Cs: Chilo suppressalis; Px: Papilio xuthus; Aga: Anopheles gambiae; Dv: Daktulosphaira vitifoliae; Dm: Drosophila melanogaster; Dp: Danaus plexippus; Tm: Tenebrio molitor; Bm: Bombyx mori; Pa: Periplaneta americana; Zn: Zootermopsis nevadensis; Api: Acyrthosiphon pisum; Sl: Spodoptera littoralis; Ls: Lasioderma serricorne; Bt: Bemisia tabaci; Ape: Antheraea pernyi: Ago: Aphis gossypii.
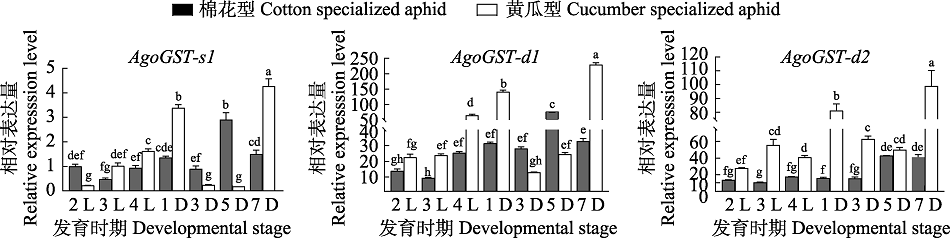
图4 AgoGST-s1、AgoGST-d1、AgoGST-d2在棉花型和黄瓜型棉蚜中的相对表达量 2 L:2龄若蚜;3 L:3龄若蚜;4 L:4龄若蚜;1 D:1 日龄成蚜;3 D:3 日龄成蚜;5 D:5 日龄成蚜;7 D:7 日龄成蚜。柱上标有不同小写字母代表基因表达量的差异显著(ANOVA, P<0.05)。
Fig. 4 Relative expression levels of AgoGST-s1, AgoGST-d1, AgoGST-d2 in cotton specialized and cucumber specialized aphids 2 L: 2nd instar nymph; 3 L: 3rd instar nymph; 4 L: 4th instar nymph; 1 D: 1st adult; 3 D: 3rd adult; 5 D: 5th adult; 7 D: 7th adult. Different lowercase letters above histograms indicate significant difference among various genes (ANOVA, P<0.05).
| [1] |
Broeders S, Huber I, Grohmann L, Berben G, Taverniers I, Mazzara M, Roosens N, Morisset D , 2014. Guidelines for validation of qualitative real-time PCR methods. Trends in Food Science & Technology, 37(2):115-126.
doi: 10.1016/j.tifs.2014.03.008 URL |
| [2] |
Cao DP, Liu Y, Walker WB, Li JH, Wang GR , 2014. Molecular Characterization of the Aphis gossypii olfactory receptor gene families. PLoS ONE, 9(6):e101187.
doi: 10.1371/journal.pone.0101187 URL pmid: 24971460 |
| [3] | Chen WS, Gu DJ, Li W, Chen ZP, Zhang WQ , 1997. Study on the host specialisation of Myzus persicae in south china. Journal of South China Agricultural University, 18(4):54-56. |
| [ 陈文胜, 古德就, 李卫, 陈泽鹏, 张维球 , 1997. 烟蚜寄主专化性的研究. 华南农业大学学报, 18(4):54-56.] | |
| [4] | Dixon DP, Edwards R , 2005. Glutathione transferases. The Arabidopsis Book, 8(45):125-131. |
| [5] |
Enayati AA, Ranson H, Hemingway J , 2005. Insect glutathione transferases and insecticide resistance. Insect Molecular Biology, 14(1):3-8.
URL pmid: 15663770 |
| [6] | Fang Y, Qiao GX, Zhang GX , 2011. Morphological adaptation of aphid species on different host plant leaves. Acta Entomologica Sinica, 54(2):157-178. |
| [ 方燕, 乔格侠, 张广学 , 2011. 不同寄主植物叶片上蚜虫的形态适应. 昆虫学报, 54(2):157-178.] | |
| [7] |
Gao F, Zhu SR, Sun YC, Du L, Parajulee M, Kang L, Ge F , 2008. Interactive effects of elevated CO2 and cotton cultivar on tri-trophic interaction of Gossypium hirsutum, Aphis gossypii and Propylaea japonica. Environmental Entomology, 37(1):29-37.
doi: 10.1603/0046-225X(2008)37[29:IEOECA]2.0.CO;2 URL pmid: 18348793 |
| [8] |
Hayaoka T, Dauterman WC , 1983. The effect of phenobarbital induction on glutathione conjugation of diazinon in susceptible and resistant house flies. Pesticide Biochemistry and Physiology, 19(3):344-349.
doi: 10.1016/0048-3575(83)90063-9 URL |
| [9] | He C, Xie W, Zhang YJ , 2017. Comparson of the responses of glutathione- S-transferase genes in Bemisia tabaci B to different host plant transfers. Plant Protection, 43(6):72-77. |
| [ 何超, 谢文, 张友军 , 2017. 谷胱甘肽-S-转移酶基因在B型烟粉虱寄主转换中的差异比较. 植物保护, 43(6):72-77.] | |
| [10] |
Hedin PA, Parrott WL, Jenkins JN , 1992. Relationships of glands, cotton square terpenoid aldehydes, and other allelochemicals to larval growth of Heliothis virescens(Lepidoptera: Noctuidae). Journal of Economic Entomology, 85(2):359-364.
doi: 10.1093/jee/85.2.359 URL |
| [11] | Jin YL, Zhang BX, Lin HF , 2018. Identifying and measuring the expression profiles, of Chilo suppressalis glutathione-S-transferase genes. Chinese Jounrnal of Applied Entomology, 55(3):349-359. |
| [ 金燕璐, 张邦贤, 林华峰 , 2018. 二化螟谷胱甘肽-S-转移酶基因的鉴定与表达模式分析. 应用昆虫学报, 55(3):349-359.] | |
| [12] |
Ketterman AJ, Saisawang C, Wongsantichon J , 2011. Insect glutathione transferases. Drug Metabolism Reviews, 43(2):253-265.
doi: 10.3109/03602532.2011.552911 URL |
| [13] |
Lee K , 1991. Glutathione- S-transferase activities in phytophagous insects: Induction and inhibition by plant phototoxins and phenols. Insect Biochemistry, 21(4):353-361.
doi: 10.1016/0020-1790(91)90001-U URL |
| [14] | Li F, Han ZJ, Wu ZF , 2002. Comparison of esterase and acetylcholinesterase of cotton aphid, Aphis gossypii Glover reared on different host plants. Journal of Nanjing Agricultural University, 25(2):57-60. |
| [ 李飞, 韩召军, 吴智锋 , 2002. 取食不同寄主棉蚜的羧酸酯酶和乙酰胆碱酯酶特性比较. 南京农业大学学报, 25(2):57-60.] | |
| [15] | Li YH , 2014. The progress in the research of insect GST on insecticide detoxification and endogenous metabolism, Journal of Shanxi Datong University (Natural Science), 30(4):49-51. |
| [ 李亚红 , 2014. 昆虫谷胱甘肽硫转移酶农药解毒与内源代谢研究进展. 山西大同大学学报(自然科学版), 30(4):49-51.] | |
| [16] |
Li ZQ, Zhang S, Luo JY, Wang CY, Lv LM, Dong SL, Cui JJ , 2013. Ecological adaption analysis of the cotton aphid ( Aphis gossypii) in different phenotypes by transcriptome comparison. PLoS ONE, 8(12):e83180.
doi: 10.1371/journal.pone.0083180 URL pmid: 24376660 |
| [17] | Liu J, Li RM, Zhou XM , 2013. Characteristics of mRNA expression of GSTs1 gene in Spodoptera exigua and the effect of chlorantraniliprole on its expression. Plant Protection, 39(3):18-21. |
| [ 刘佳, 李如美, 周小毛 , 2013. 甜菜夜蛾GSTs1基因mRNA表达特征及氯虫苯甲酰胺对其表达量的影响. 植物保护, 39(3):18-21.] | |
| [18] |
Liu S, Zhang YX, Wang WL, Zhang BX, Li SG , 2017. Identification and characterisation of seventeen glutathione- S-transferase genes from the cabbage white butterfly Pieris rapae. Pesticide Biochemistry and Physiology, 143:102-110.
doi: 10.1016/j.pestbp.2017.09.001 URL pmid: 29183577 |
| [19] | Liu XD, Zhai BP, Zhang XX , 2003. Studies on the host biotypes and its cause of cotton aphid in Nanjing, China. Scientia Agricultura Sinica, 36(1):54-58. |
| [ 刘向东, 翟保平, 张孝羲 , 2003. 南京地区棉蚜寄主专化型及其成因研究. 中国农业科学, 36(1):54-58.] | |
| [20] | Liu XD, Zhang XX, Zhai BP , 2004. Host biotypes and their formation causes in aphids. Acta Entomologica Sinica, 47(4):499-506. |
| [ 刘向东, 张孝羲, 翟保平 , 2004. 蚜虫寄主专化型及其成因. 昆虫学报, 47(4):499-506.] | |
| [21] | Liu ZH, Zhao GH, Lu JS, Shui Y, Wu G , 2008. Studies on content of cotton gossypol and characteristics of pest resistance. Xinjiang Agricultural Sciences, 45(3):409-413. |
| [ 刘泽辉, 赵国虎, 陆敬善, 水涌, 武刚 , 2008. 棉花棉酚含量与抗虫特性的研究. 新疆农业科学, 45(3):409-413.] | |
| [22] |
Livak KJ, Schmittgen TD , 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2 -ΔΔCT method . Methods, 25(4):402-408.
doi: 10.1006/meth.2001.1262 URL pmid: 11846609 |
| [23] |
Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ, Huang YP, Chen XY , 2007. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nature Biotechnology, 25(11):1307-1313.
doi: 10.1038/nbt1352 URL pmid: 17982444 |
| [24] |
Masutti FV, Chavigny P , 1998. Host-based genetic differentiation in the aphid Aphis gossypii Glover, evidenced from RAPD fingerprints. Molecular Ecology, 7(7):905-914.
doi: 10.1046/j.1365-294x.1998.00421.x URL |
| [25] |
Masutti FV, Chavigny P, Fuller SJ , 1999. Characterization of microsatellite loci in the aphid species Aphis gossypii Glover. Molecular Ecology, 8(4):693-695.
doi: 10.1046/j.1365-294x.1999.00876.x URL pmid: 10327667 |
| [26] |
Ranson H, Claudianos C, Ortelli F, Abgrall C, Hemingway J, Sharakhova MV, Unger MF, Collins FH, Feyereisen R , 2002. Evolution of supergene families associated with insecticide resistance. Science, 298(5591):179-181.
doi: 10.1126/science.1076781 URL pmid: 12364796 |
| [27] |
Ranson H, Rossiter L, Ortelli F, Jensen B, Wang Xl, Roth CW, Collins FH, Hemingway J , 2001. Identification of a novel class of insect glutathione S-transferases involved in resistance to DDT in the malaria vector Anopheles gambiae. Biochemical Journal, 359(2):295-304.
doi: 10.1042/bj3590295 URL |
| [28] |
Reidy GF, Rose HA, Visetson S, Murray M , 1990. Increased glutathione- S-transferase activity and glutathione content in an insecticide-resistant strain of Tribolium castaneum(Herbst). Pesticide Biochemistry and Physiology, 36(3):269-276.
doi: 10.1016/0048-3575(90)90035-Z URL |
| [29] | Ren KY, Zhang S, Luo JY, Wang CY, Lv LM, Zhang LJ, Zhu XZ, Wang L, Cui JJ, Zhu JB , 2018. Clone and expression analysis of salivary gland protein C002 in different host-specific types of Aphis gossypii Glover. Journal of Environmental Entomology, 40(3):617-623. |
| [ 任柯昱, 张帅, 雒珺瑜, 王春义, 吕丽敏, 张利娟, 朱香镇, 王丽, 崔金杰, 祝建波 , 2018. 棉蚜唾液蛋白C002基因克隆及在棉蚜不同寄主专化型中的表达分析. 环境昆虫学报, 40(3):617-623.] | |
| [30] |
Saito T , 1990. Insecticide resistance of the cotton aphid, Aphis gossypii Glover (Homoptera: Aphididae): Variation of esterase activity by host plants. Japanese Journal of Applied Entomology and Zoology, 34(1):37-41.
doi: 10.1303/jjaez.34.37 URL |
| [31] | Tang DL , 1999. The secondary metabolism of plant insect resistant. World Agriculture, 239(3):32-33. |
| [ 汤德良 , 1999. 植物抗虫的次生代谢物质. 世界农业, 239(3):32-33.] | |
| [32] | Tang F, Liang P, Gao XW , 2005. 2-tridecylketone and quercetin induce tissue-specific expression of glutathione- S-transferase in Helicoverpa armigera. Natural Science Exhibition, 15(7):805-810. |
| [ 汤方, 梁沛, 高希武 , 2005. 2-十三烷酮和槲皮素诱导棉铃虫谷胱甘肽-S-转移酶组织特异性表达. 自然科学进展, 15(7):805-810.] | |
| [33] |
Vontas JG, Small GJ, Hemingway J , 2001. Glutathione- S-transferases as antioxidant defence agents confer pyrethroid resistance in Nilaparvata lugens. Biochemical Journal, 357(1):65-72.
doi: 10.1042/bj3570065 URL |
| [34] |
Wadleigh RW, Yu SJ , 1987. Glutathione transferase activity of fall armyworm larvae toward α, β-unsaturated carbonyl allelochemicals and its induction by allelochemicals. Insect Biochemistry, 17(5):759-764.
doi: 10.1016/0020-1790(87)90046-1 URL |
| [35] |
Wadleigh RW, Yu SJ , 1988. Detoxification of isothiocyanate allelochemicals by gluthione- S-transferase in three lepidopterous species. Journal of Chemistry Ecology, 14(5):1279-1288.
doi: 10.1007/BF01019352 URL |
| [36] | Wang L , 2015. Preliminary studies on the host biotypes and formation mechanism of cotton aphid in cotton areas of north China. Master dissertation. Anyang: Chinese Academy of Agricultural Sciences. |
| [ 王丽 , 2015. 华北棉区棉蚜寄主专化型及其形成机制初步研究. 硕士学位论文. 安阳: 中国农业科学院.] | |
| [37] | Wang L, Zhang S, Luo JY, Wang CY, Lü LM, Zhu XZ, Li CH, Cui JJ , 2016. Identification of Aphis gossypii Glover (Hemiptera: Aphididae) biotypes from different host plants in north China. PLoS ONE, 11(1):34-39. |
| [38] | Wang SY, Zhou XH, Zhang AS, Li LL, Men XY, Liu YJ, Li XK, Yu Y , 2013. Mechanisms and cross-resistance of imidacloprid resistance in Frankliniella occidentalis. Chinese Jounrnal of Applied Entomology, 50(1):167-172. |
| [ 王圣印, 周仙红, 张安盛, 李丽莉, 门兴元, 刘永杰, 李许可, 于毅 , 2013. 西花蓟马对吡虫啉的抗性机制及交互抗性研究. 应用昆虫学报, 50(1):167-172.] | |
| [39] | Wu YQ, Guo YY, Yang J , 2000. Analysis of flavonoid substance in cotton plants for resistance to pests by HPLC. Plant Protection, 26(5):1-3. |
| [ 武予清, 郭予元, 杨舰 , 2000. 棉株中抗虫物质黄酮类化合物的高效液相色谱分析. 植物保护, 26(5):1-3.] | |
| [40] |
Yang J, Sun XQ, Yan SY, Pan WJ, Zhang MX, Ca QN , 2017. Interaction of ferulic acid with glutathione- S-transferase and carboxylesterase genes in the brown planthopper, Nilaparvata lugens. Journal of Chemical Ecology, 43(7):1-10.
doi: 10.1007/s10886-016-0812-x URL |
| [41] | You YC, Xie M, You MS , 2013. The diversity and role of glutathione- S-transferase in insecticide resistance in insects. Chinese Journal of Applied Entomology, 50(3):831-840. |
| [ 尤燕春, 谢苗, 尤民生 , 2013. 昆虫谷胱甘肽-S-转移酶的多样性及其介导的抗药性. 应用昆虫学报, 50(3):831-840.] | |
| [42] |
Yu SJ , 1982. Host plant induction of glutathione- S-transferase in the fall armyworm. Pesticide Biochemistry and Physiology, 18(1):101-106.
doi: 10.1016/0048-3575(82)90092-X URL |
| [43] |
Yu SJ , 1984. Interactions of allelochemicals with detoxication enzymes of insecticide-susceptible and resistant fall armyworms [ Spodoptera frugiperda, host plants]. Pesticide Biochemistry and Physiology, 22(1):60-68.
doi: 10.1016/0048-3575(84)90010-5 URL |
| [44] |
Yu SJ , 1999. Induction of new glutathione- S-transferase isozymes by allelochemicals in the fall armyworm. Pesticide Biochemistry and Physiology, 63(3):163-171.
doi: 10.1006/pest.1999.2402 URL |
| [45] | Yuan LB, Gao ZL, Dang ZH, Li YF, Chi GT, Pan WL , 2008. Fitness differentiation in Aphids gossypii populations to cotton and cucumber. Chinese Bulletin of Entomolog, 45(6):896-899. |
| [ 袁立兵, 高占林, 党志红, 李耀发, 赤国彤, 潘文亮 , 2008. 不同棉蚜种群对棉花和黄瓜的适合度分化. 昆虫知识, 45(6):896-899.] | |
| [46] | Zhai YT , 2015. Defense function analysis of three host secondary substances for different biotypes of Sitobion avenae. Master dissertation. Xianyang: Northwest A&F University. |
| [ 翟颖婷 , 2019. 三种寄主次生物质对不同生物型麦长管蚜的防御功能分析. 硕士学位论文. 咸阳:西北农林科技大学.] | |
| [47] |
Zhang S, Luo JY, Wang L, Wang CY, Lv LM, Zhang LJ, Zhu XZ, Cui JJ , 2018. The biotypes and host shifts of cotton-melon aphis Aphis gossypii in northern China. Journal of Integrative Agriculture, 17(9):2066-2073.
doi: 10.1016/S2095-3119(17)61817-3 URL |
| [48] | Zhang YC , 2016. The regulation mechanisms of population density of Buchnera aphidicola in Aphis gossypii Glover. Doctoral dissertation. Nanjing: Nanjing Agricultural University. |
| [ 张元臣 , 2016. 棉蚜体内原生共生菌Buchnera aphidicola种群密度的调控机制. 博士学位论文. 南京: 南京农业大学.] | |
| [49] | Zhu JY , 2017. Host switching affects the fitness and metabolism of Myzus persicae. Master dissertation. Xianyang:Northwest A&F University. |
| [ 朱经云 , 2017. 寄主转换对桃蚜适合度及生理代谢的影响. 硕士学位论文. 咸阳: 西北农林科技大学.] |
| No related articles found! |
| 阅读次数 | ||||||
|
全文 |
|
|||||
|
摘要 |
|
|||||
